firestar [1,2] EXTENDED Results Output
Target sequences are subjected to standard HHsearch[3] and PSI-BLAST[4] searches. Profiles are generated with an nrdb70 database from the EBI and the final search is made against the FireDB[5] consensus sequence database derived from PDB sequences in the case of PSI-BLAST, and against an ad hoc profile database generated starting from the previous one in the case of HHsearch.
Consensus sequences are identified by id's of the kind t_1tcoC where the chain "C" of the pdb file "1tco" is the representative of the cluster. Consensus sequences are built from clusters of sequences with a similarity of 97% sequence identity.E-values can be modified by the user. The default E-value cutoff is set to 10. We have found [6] that in some cases local similarities are reliable enough to transfer binding site residues between even distant homologues.
Only those PSI-BLAST and HHsearch hits with any functional information annotated in FireDB are displayed. Each hit is represented in its own box. All boxes are displayed on the same page.
Each alignment is evaluated using a specially adapted version of SQUARE [7]. This method generates scores for each position in the alignment based on local conservation scores.
SQUARE produces reliability scores for all aligned residues, but it is important to note that these reliability scores refer solely to the viability of the transference of functionally important residues from the consensus sequence. The score is meaningless for those residues in the FireDB consensus sequence that do not have associated functionally important residues.
The functionally important residues from the FireDB consensus sequences are supported by evidence. In the case of catalytic residues the evidence comes from the Catalytic Site Atlas [8] and are annotated as "literature" or "inferred with PSI-BLAST". In the case of binding residues a percentage of occurrence is generated. The following table explains evidence and reliability scores in more detail:
| SQUARE: table of scores | Binding site occurrence and catalytic residue evidence | |||
| - | Gapped or non-conserved position | literature | Catalytic residue annotated from literature | |
| 1 | 45% reliability | PSI-BLAST | Catalytic residue inferred by similarity | |
| 2 | 60% reliability | |||
| 3 | 75% reliability | 0-20% occurrence | ||
| 4 | 85% reliability | 20-50% occurrence | ||
| 5 | 90% reliability | 50-100% occurrence | ||
| C | 99% reliability | XX% occurrence | Not applicable | |
| Note that sites are usually formed by groups of residues. The user should assess overall conservation based on all the residues important for one site. It is common to find that binding sites are partially conserved. | Occurrence is calculated when a given consensus sequence has several representatives in the PDB. It indicates the percentage of PDB representatives that bind ligand analogs at same site. Where there are less than 5 representatives the values are marked "not applicable". | |||
PSI-BLAST and HHsearch output guide scheme
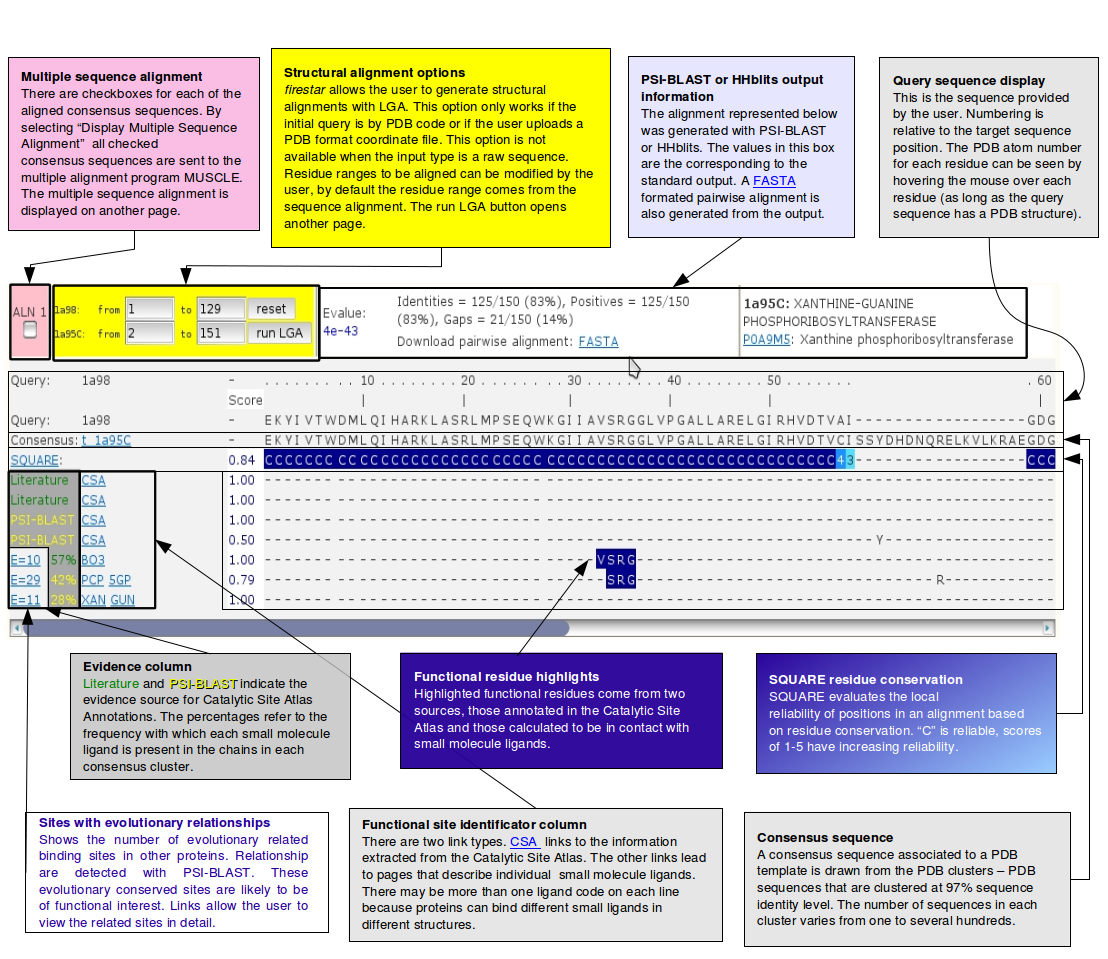
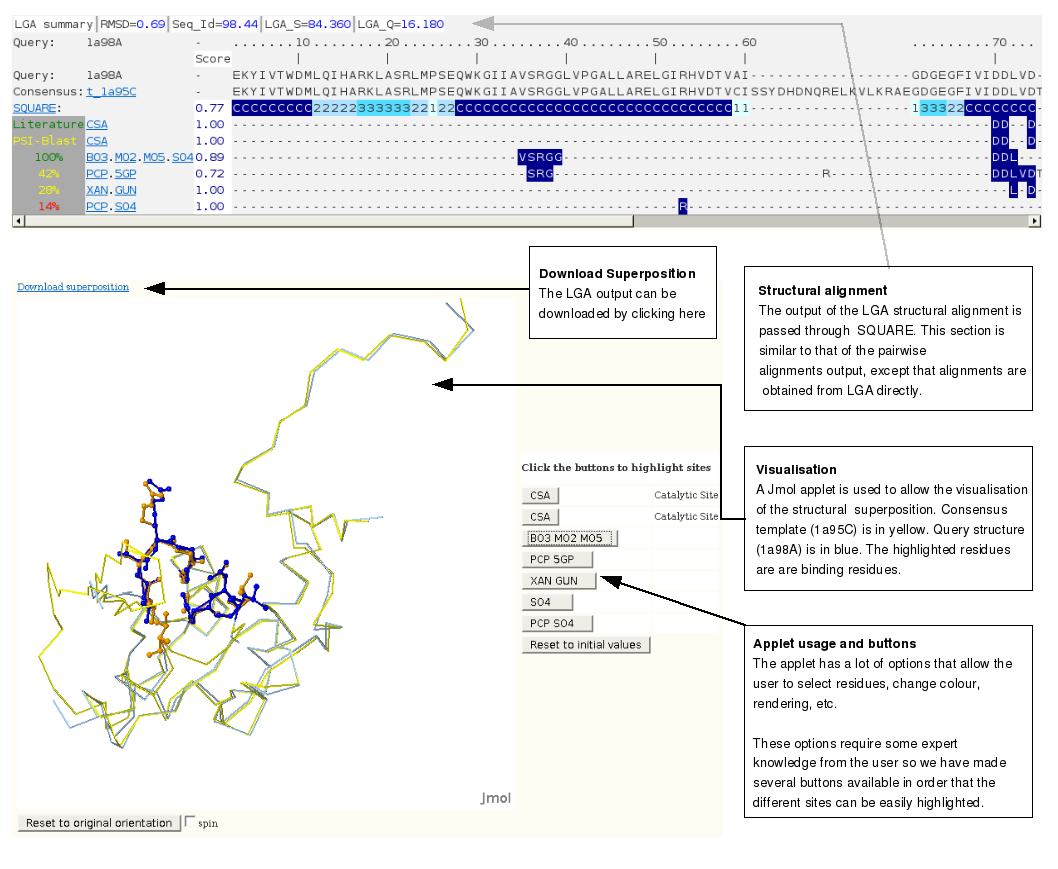
Structural alignment outputStructural alignments can be generated by clicking on "run LGA" in the yellow box next to each PSI-BLAST and HHsearch output. Structural superpositions between the query and the representative of the selected cluster are generated with LGA [9] and open in a separate page that has a similar look to the pairwise results. The superpositions can be visualised within the page with a Jmol applet. Note that this means that Java environment must be installed for the navigator. If the java environment is not installed for the browser the applet may cause the navigator to crash on some platforms. Unfortunately this technical issue is beyond our control. |
Structural alignment output guide scheme
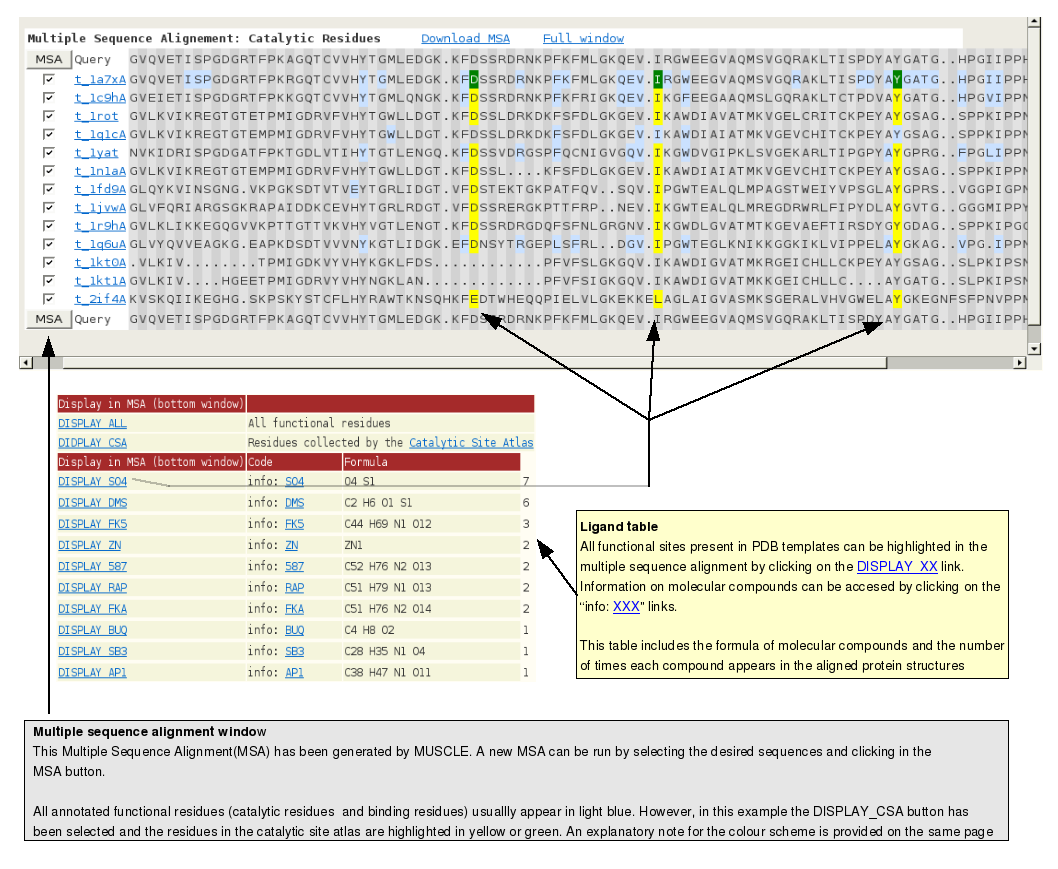
Multiple sequence alignment outputMultiple alignments between the consensus sequences and the query sequence can be generated by clicking on the "Display Multiple Sequence" button in
the pink box in the PSI-BLAST or HHsearch output page. MUSCLE [10] will align the query sequence to all the consensus sequences found by the two programs.
| |||||||||||||||||||
Multiple sequence alignment output guide scheme
firecat help pageAlignment reliability is most sensitive to alignment quality. This tool allows user to insert their own pairwise query-template alignments with the requirement that template must be a PDB sequence. It requires the second sequence in the pairwise alignment to be a PDB sequence. In this way the server can identify the FireDB cluster to which the second sequence belongs and associate the functional information stored in FireDB. This tool can be used when alignments can be improved, but also when firestar PSI-BLAST and HHsearch are unable to find homologues. In this case query-template alignments from fold recognition servers can be used. exampleThe following alignment has been generated by the FFAS03 [11] threading server and was the best scoring alignment for target T0369 of the CASP7 experiment in the 3D-Jury [12] metaserver. The structural template identified by FFAS was 1rxq_A and we can assess whether the functional residues transfer between the target and the template. >T0369 MTDWQQALDRHVGVGVRTTRDLIRLIQPEDWDKRPISGKRSVYEVAVHLAVLLEADLRIATG-------- -----------ATADEMAQFYAVPVLPEQLVDRLDQSWQYYQDRLMADFSTETTYWGVTDSTTGWLLEAA VHLYHHRSQLLDYLNLLGYDIKLDLFE >1rxq_A SKEQKDKWIQVLEEVPAKLKQAVEVMTDSQLDTPYRDGGWTVRQVVHHLADSHMNSYIRFKLSLTEETPA IRPYDEKAWSELKDSKTADPSGSLALLQELHGRWTALLRTLTDQQFKRGFYHPD-TKEIITLENALGLYV WHSHHHIAHITELSRRMGWS------- We paste the alignment in the text box, with the template PDB code: "1rxq" and the chain identifier: "A". And the firecat output is: |
| Query: | T0369 | . | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | 60 | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | 120 | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | |||||||||||||||||||||||||||||||||||||||
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Query: | T0369 | M | T | D | W | Q | Q | A | L | D | R | H | V | G | V | G | V | R | T | T | R | D | L | I | R | L | I | Q | P | E | D | W | D | K | R | P | I | S | G | K | R | S | V | Y | E | V | A | V | H | L | A | V | L | L | E | A | D | L | R | I | A | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | T | A | D | E | M | A | Q | F | Y | A | V | P | V | L | P | E | Q | L | V | D | R | L | D | Q | S | W | Q | Y | Y | Q | D | R | L | M | A | D | F | S | T | E | T | T | Y | W | G | V | T | D | S | T | T | G | W | L | L | E | A | A | V | H | L | Y | H | H | R | S | Q | L | L | D | Y | L | N | L | L | G | Y | D | I | K | L | D | L | F | E | |
| Template: | 1rxqB | S | K | E | Q | K | D | K | W | I | Q | V | L | E | E | V | P | A | K | L | K | Q | A | V | E | V | M | T | D | S | Q | L | D | T | P | Y | R | D | G | G | W | T | V | R | Q | V | V | H | H | L | A | D | S | H | M | N | S | Y | I | R | F | K | L | S | L | T | E | E | T | P | A | I | R | P | Y | D | E | K | A | W | S | E | L | K | D | S | K | T | A | D | P | S | G | S | L | A | L | L | Q | E | L | H | G | R | W | T | A | L | L | R | T | L | T | D | Q | Q | F | K | R | G | F | Y | H | P | D | - | T | K | E | I | I | T | L | E | N | A | L | G | L | Y | V | W | H | S | H | H | H | I | A | H | I | T | E | L | S | R | R | M | G | W | S | - | - | - | - | - | - | - | |
| Score: | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2 | 1 | 1 | - | - | - | 1 | - | - | 1 | 3 | 2 | 2 | 3 | 2 | 2 | 5 | 3 | 3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | 1 | 1 | 2 | - | - | 2 | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 2 | 2 | - | - | - | 2 | 2 | 3 | 1 | 2 | 3 | 5 | 2 | 2 | 1 | 1 | - | - | - | - | - | - | - | 2 | 1 | - | - | - | - | - | - | - | - | ||
| 100% | NI | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | - | - | - | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
|
Based on this output, the nickel binding site can be reliably transferred from 1rxq_A to the target T0369. Note also that 1rxqA could not be obtained by standard PSI-BLAST searches. |
Evolutionary relationships between sitesA binding site is deemed to be of biological relevance if it can be shown to have been conserved. The sites shown on this page are pre-calculated from data in FireDB, are highly conserved and are therefore highly likely to be be evolutionarily related. | |
Evolutionary relationships guide scheme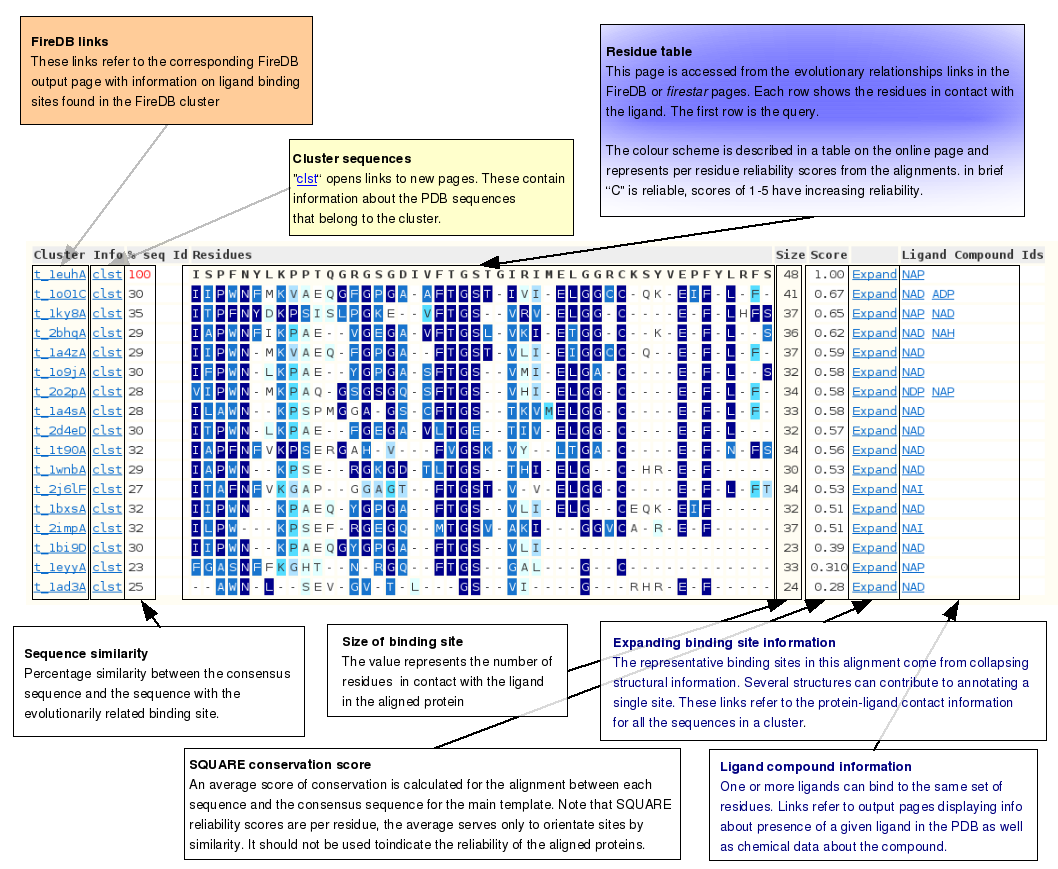
|